Multiplex-detection for Biosensing
Single-Molecule Sensors
Quantitative and simultaneous detection of biomarkers
Virginia Commonwealth University researchers have developed a novel multiplexed method for quantitative detection of nucleic acid sequences. Current methods rely on sophisticated engineered solutions such as microarrays, DNA barcodes, and synthetic nanopores. While effective, these methods can only provide semi-quantitative information. Although single-molecule FRET (smFRET) microscopy has become an attractive tool for biomarker analysis, as it allows for sensitive and quantitative detection, utilizing this tool remains a challenge as it is only able to analyze one target at a time and multiplexing with this technique traditionally requires complex labeling schemes and data analysis algorithms.
Our researchers have eliminated the need for these complex systems by developing a method which employs a series of interconvertible Hairpin-based Sensors (iHabSs). These iHabSs are able to achieve non-overlapping FRET signals which allows for simultaneous detection of multiple unlabeled nucleic acids.
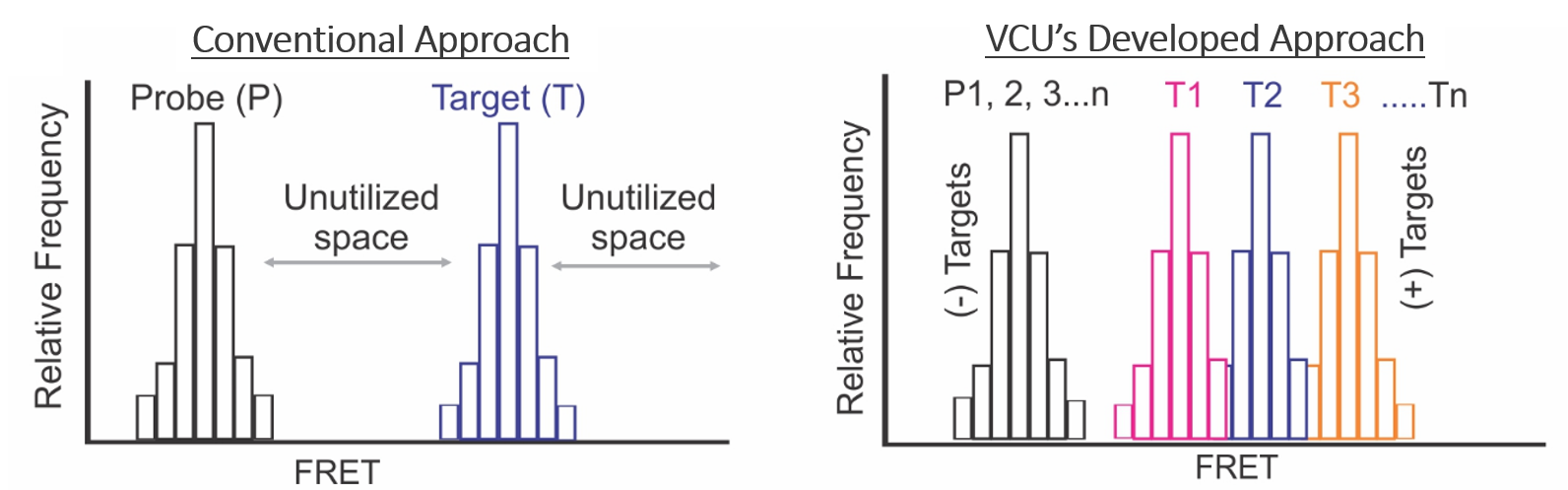
Figure 1: Left: conventional approach that allows the detection of only one target at a time. Right: Our approach for simultaneous detection of multiple targets by filling the unutilized FRET spaces.
The technology
The iHabSs are comprised of a DNA-hairpin that is flanked on both sides by thymine spacers. This structure is then sandwiched by two short pieces of double-stranded DNA, each labeled with a donor or acceptor fluorophore. Multiplexing is achieved by using a combination of these iHabSs with various lengths of flanking thymine spacers as seen below in Figure 2a.
While existing antenna systems require multiple FRET pairs, this approach uses only one. The novel design of the DNA hairpin sensor allows for the detection of a target due to an increase in the FRET, as seen in Figure 2b. By using a combination of DNA hairpins, this method is able to utilize the gaps between high and low FRET signals to provide simultaneous information on multiple targets. These iHabSs are highly sensitive, allowing for the detection of picomolar (pM) concentrations of unlabeled nucleic acid targets and bear tremendous promise for quantitative detection of unmodified targets in their native environments. Additionally, these sensors are designed to be fully recyclable for multiple rounds of detection and cost-effective as they can be readily prepared from short synthetic DNA strands.
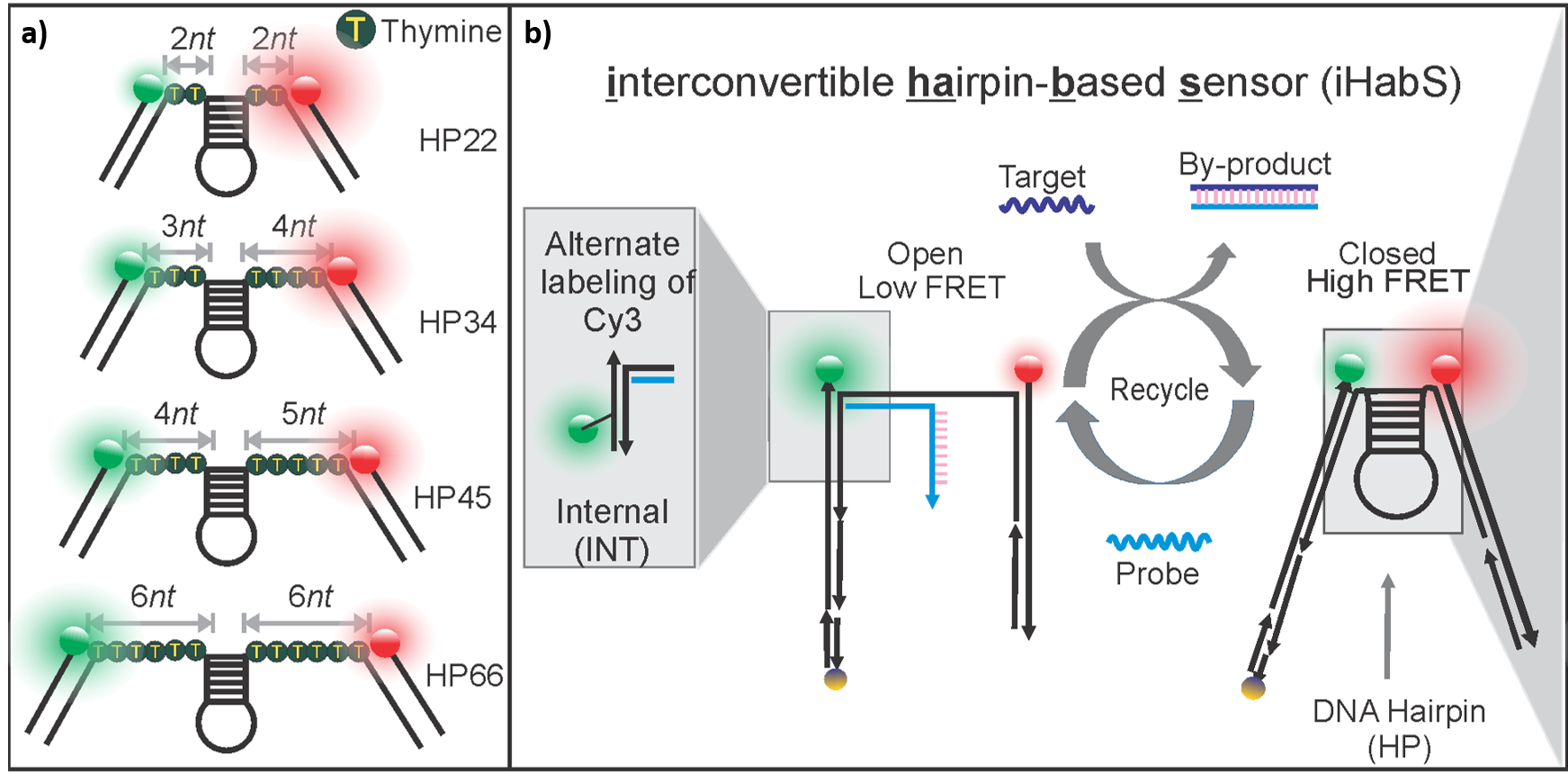
Figure 2: (a) Hairpins (HP) with various flanking thymine spacers (represented as the number of thymine nucleotides “nt”) used to tune the FRET; (b) Working principle of iHabSs.
